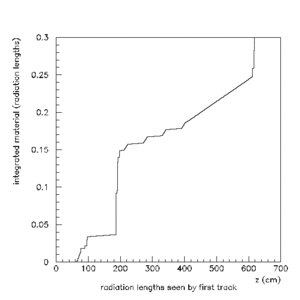
HOWTO do a Radiation Length Scan
Mechanisms have been built into hdgeant to allow one to use it to produce radiation length scans of the geometry defined in the program. There are a couple of ways to do this. One uses the hbook file produced by hdgeant itself while the other uses DANA to produce a ROOT file. Both are described here.
Setting up
First, you need to make sure you have the geometry you want to scan defined in hdgeant. For info on how to do this, see "Modifying the geometry".
You will also want to limit the step size Geant uses while tracking. By default, Geant automatically calculates its step sizes for each material [1]. The problem with this is that low density materials will often be stepped over in one large step, making the material appear as though it is concentrated at the end of the step rather than dispersed along the path. To turn off the use of the automatic step size calculator globally, one needs to set AUTO=0 in the control.in file.
The default step size set for materials when AUTO is off is 1 cm. This default is assigned in the hdds-geant [2] program that generates the hddsGeant3.F file from the hdds source document. The value is set using the STEMAX variable. This value can be reduced to obtain more fine-grained profiling of the geometry materials either by editing a private copy of hdds-geant.cpp and using it to generate a custom hddsGeant3.F file from the hdds source document, or simply by doing a universal search-and-replace in hddsGeant3.F using a text editor and using it to build a custom hdgeant executable. Note that such a modified executable should not be used to carry out physics simulations; running Geant3 with non-optimized tracking parameters requires special care to obtain correct simulation results.
You should also use geantinos(GEANT id = 48) or at least turn off multiple scattering and energy loss in your control.in file using the MULS and LOSS keys. Here is an example of a simple control.in file useful for this purpose:
TRIG 1 RUNG 9999 OUTFILE 'hdgeant.hddm' c The following card enables single-track generation (for testing). c For a single-particle gun, set the momentum (GeV/c), direction c theta,phi (degrees) and vertex position (cm), and for the particle c type insert the Geant particle type code plus 100 (eg. 101=gamma, c 103=electron, 107=pi0, 108=pi+, 109=pi-, 114=proton). If you use c the particle code but do not add 100 then theta,phi are ignored c and the particle direction is generated randomly over 4pi sr. c For a listing of the Geant particle types, see the following URL. c http://wwwasdoc.web.cern.ch/wwwasdoc/geant_html3/node72.html c The meaning of the arguments to KINE are as follows. c - particle = GEANT particle type of primary track + 100 c - momentum = initial track momentum, central value (GeV/c) c - theta = initial track polar angle, central value (degrees) c - phi = initial track azimuthal angle, central value (degrees) c - delta_momentum = spread in initial track momentum, full width (GeV/c) c - delta_theta = spread in initial track polar angle, full width (degrees) c - delta_phi = spread in initial track azimuthal angle, full width (degrees) c c particle momentum theta phi delta_momentum delta_theta delta_phi KINE 148 3.1 3. 180. 0. 0. 360. c The SCAP card determines the vertex position for the particle gun. It c supports the following three arguments, all of which default to 0. c c vertex_x vertex_y vertex_z SCAP 0. 0. 65. RNDM 172 CUTS 1e-4 1e-4 SWIT 0 0 0 0 0 0 0 0 0 0 C The NOSECONDARIES card is specific to HDGeant. Setting it to a non-zero C integer will cause the calling of GSKING() to be suppress leaving C perfectly clean tracks NOSECONDARIES 1 c TRAJECTORIES = 0 don't store trajectory info c TRAJECTORIES = 1 store birth and death points of primary tracks c TRAJECTORIES = 2 store birth and death points of all particles c TRAJECTORIES = 3 store full trajectory of primary tracks c TRAJECTORIES = 4 store full trajectory of primary tracks and birth/death points of secondaries c TRAJECTORIES = 5 store full trajectory for all particles c TRAJECTORIES 3 AUTO 0 END
HBOOK / PAW method
With this method, you can get a couple of historgrams from hdgeant directly containing the amount of material in g/cm2 and the number of radiation lengths seen by the first track. To do this, you need to add the following line to the top of the gustep.F file in the HDGeant directory and re-compile:
#define HISTOGRAM_MATERIAL_SEEN_BY_FIRST_TRACK 1
Run hdgeant and a file called "geant.hbook" will be produced. Open it with paw like so:
>paw Calling PRO version of paw-X11 ****************************************************** * * * W E L C O M E to P A W * * * * Version 2.14/04 12 January 2004 * * * ****************************************************** Workstation type (?=HELP) <CR>=1 : Version 1.29/04 of HIGZ started *** No default PAWLOGON file "/Users/davidl/.pawlogon.kumac" found PAW > hi/fil 0 geant.hbook 65536 PAW > hi/li Note that the histograms are not integrated material so they must be integrated "by hand" to find the total material seen by a track.
===> Directory :
1 (1) material in g/cm2 seen by first track
1000001 (1) radiation lengths seen by first track
PAW > v/cre tmp(1000)
PAW > vect/get 1000001 tmp
PAW > sigma tmp=sum(tmp)
PAW > vect/put 1000001 tmp
PAW > max 1000001 0.3
PAW > hi/pl 1000001
PAW > atit 'z (cm)' 'integrated material (radiation lengths)'
A series of scans done using the HBOOK/PAW method can be found here.
DANA/ROOT method
With the DANA/ROOT method, one can incorporate multiple tracks to scan the average amount of material seen for a range of angles. If you wish to do this, change the TRIG value to something larger and modify the KINE keys appropriately.
Run hdgeant and then run hd_root with the radlen_hists plugin on the resulting hdgeant.hddm file like this:
hd_root --plugin=radlen_hists hdgeant.hddm
This will produce a file called "hd_root.root" that contans some histograms. To open it and look at the histogram of integrated radiation lengths as a function of z, do the following:
>root hd_root.root ******************************************* * * * W E L C O M E to R O O T * * * * Version 5.14/00 14 December 2006 * * * * You are welcome to visit our Web site * * http://root.cern.ch * * * ******************************************* FreeType Engine v2.1.9 used to render TrueType fonts. Compiled on 15 December 2006 for macosx with thread support. CINT/ROOT C/C++ Interpreter version 5.16.16, November 24, 2006 Type ? for help. Commands must be C++ statements. Enclose multiple statements between { }. root [0] Attaching file hd_root.root as _file0... root [1] .ls TFile** hd_root.root Produced by hd_root TFile* hd_root.root Produced by hd_root KEY: TDirectory RADLEN;1 RADLEN KEY: TTree ROOTtree;1 HDGeant Hits Tree root [2] RADLEN.cd() (Bool_t)1 root [3] .ls TDirectory* RADLEN RADLEN KEY: TH2F dE_vs_r;1 dE (keV) vs r KEY: TH2F dE_vs_z;1 dE (keV) vs z KEY: TH1F theta_nevents;1 Number of events per #theta bin in int_radlen_vs_z_vs_theta KEY: TH2F nXo_vs_z_vs_theta;1 Radiation lengths vs. z and #theta KEY: TH2F inXo_vs_z_vs_theta;1 Integrated radiation lengths vs. z and #theta KEY: TH2F nXo_vs_r_vs_theta;1 Radiation lengths vs. r and #theta KEY: TH2F inXo_vs_r_vs_theta;1 Integrated radiation lengths vs. r and #theta KEY: TH1F nXo_vs_z;1 Radiation lengths vs. z KEY: TH1F inXo_vs_z;1 Integrated radiation lengths vs. z KEY: TH1F nXo_vs_r;1 Radiation lengths vs. r KEY: TH1F inXo_vs_r;1 Integrated radiation lengths vs. r root [4] inXo_vs_z->Draw() <TCanvas::MakeDefCanvas>: created default TCanvas with name c1 root [5]