CDC algo status
From GlueXWiki
Add simplifications to offline code to show impact
Using prototype data from CDC_50_50 (run 31942) with offline analysis
50/50 Ar/CO2 and cosmics, 2100V, prototype horizontal
Only 20 samples upsampled
- Event pedestal is mean of 100 samples ending at trigger time
- Find hit threshold crossing
- Upsample 20 samples starting 10 before hit threshold crossing
- Find hit threshold crossing again in upsampled data
- Step back <pedlead> points to find new local pedestal
- Search forward to find high threshold crossing
- Search backward to find low threshold crossing
- Project through both thresholds to find pedestal crossing time
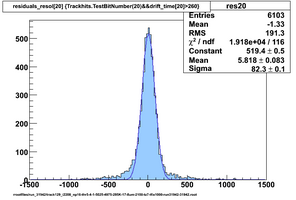
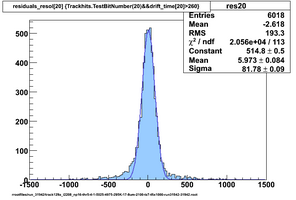
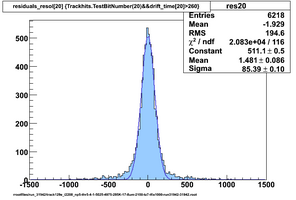
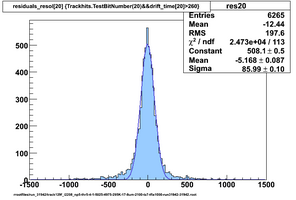
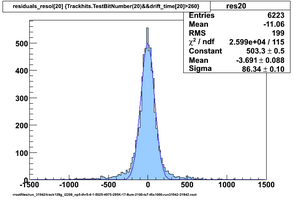
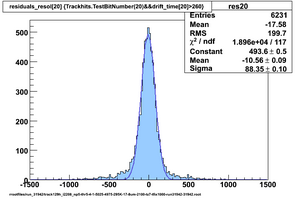
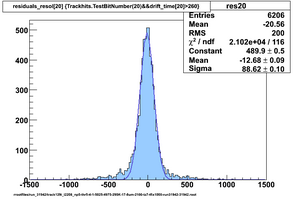
Selected results below
Lower line (c and d) are from single threshold crossings