GlueX Offline FAQ
This is the GlueX Offline Frequently-Asked Questions list. If you cannot find an answer to your question, please send it to the Offline Software Coordinator (currently Mark Ito).
Contents
- 1 General
- 2 GlueX Administrative Tasks
- 2.1 How do I get an account on the wiki?
- 2.2 How do I get privilege to check stuff into the Subversion repository?
- 2.3 How do I get emails when files are checked into the Subversion repository? Stop the emails from coming?
- 2.4 How do I add a new author to the DocDB?
- 2.5 How do I get write access to the GlueX GitHub repositories?
- 2.6 How do join the JLab Slack channel?
- 3 Building GlueX Software
- 4 JLab
- 4.1 How do I get a computer account at JLab?
- 4.2 How do I install a certificate in my browser so that I can view secure web sites at JLab (without creating a security exception)?
- 4.3 I cannot log into the ifarm machines. How do I get access?
- 4.4 How do I become a member of the "halld" Unix group?
- 4.5 How do I subscribe to a GlueX email list?
- 4.6 How do I get personal web space at JLab?
- 4.7 What are the various types of public disk space available at JLab?
- 4.8 What is the scratch disk?
- 4.9 What is the Volatile disk?
- 4.10 What is the Cache disk?
- 4.11 How do I check how much space Hall D is using on the Lustre disks?
- 4.12 Why does sudo not work for me?
- 4.13 How do I kill all of my farm jobs?
- 4.14 How to access RCDB with python (e.g. plot accumulated triggers)
- 4.15 How do I get a Scientific Computing Certificate?
- 4.16 How do I find out what share of the Farm is allocated to Hall D?
- 4.17 How do I run a remote desktop session on a JLab machine from my Windows machine?
- 4.18 How do I connect to the JLab VPN?
- 4.19 Is there a way to comment on a ServiceNow incident opened by another user?
- 4.20 Farm Jobs
- 4.20.1 How do farm jobs handle input files requested in the job specification?
- 4.20.2 What is the relationship between slurm jobs and auger jobs?
- 4.20.3 Where do I find a list of valid PROJECT fields for farm jobs?
- 4.20.4 Is there an Auger/Slurm FAQ page somewhere?
- 4.20.5 What is this sbatch error that I am getting?
- 4.21 SWIF
- 4.22 JLab Administrative Topics
- 5 halld_recon
- 6 halld_sim
- 7 CCDB
- 8 Simulation
- 9 Git
- 9.1 Where can I get help with Git?
- 9.2 What are the basic git commands that I need to know?
- 9.3 What happens when I do a git clone?
- 9.4 What is a remote repository?
- 9.5 What is a tracking branch?
- 9.6 How do I create a tracking branch?
- 9.7 How do I make an existing branch track a remote branch?
- 9.8 What is a topic branch?
- 9.9 What is the difference between Git and GitHub?
- 9.10 How do I get privilege to create pull requests on GitHub?
- 9.11 How do I authenticate to GitHub from the command line?
- 9.12 Why is there no "git pull-request" command?
- 9.13 How are we notified of pull requests?
- 9.14 What other notifications are available?
- 9.15 What is the difference between a clone and a fork?
- 9.16 What happens to forks when a repository is deleted or changes visibility?
- 9.17 What happens when I checkout another branch when there are uncommitted changes on the current branch?
- 9.18 I cannot clone a GitHub repository on the Gluon Cluster. What is the trick?
- 9.19 I am getting a "403 Forbidden" error when trying to push to a GitHub repository from JLab. What is wrong?
- 9.20 After a branch is deleted on GitHub, I still see it when I do a git branch -r on my local repository, even after a git fetch. How do I get rid of them?
- 9.21 How do I get to the compare view on GitHub?
- 9.22 Tags have changed on a remote repository. How do I update them on my local repository?
- 9.23 I am getting SSL errors when trying to access GitHub from JLab. What is wrong?
- 9.24 How do I use secure shell (ssh) to access GitHub?
- 9.25 How do I restart the automatic pull-request test?
- 9.26 How do I find the GlueX-related repositories on GitHub?
- 9.27 How do I get notified of changes in a repository on GitHub?
- 10 ROOT
General
Where do I find information about GlueX software?
The best place is this wiki. The highest-level page dealing with software is the Offline Software page. The offline page is linked from the front page of the wiki at http://wiki.gluex.org .
GlueX Administrative Tasks
How do I get an account on the wiki?
All members of the halld group at JLab can log into the wiki. Use your JLab CUE username and password.
How do I get privilege to check stuff into the Subversion repository?
Become a member of the halld unix group.
How do I get emails when files are checked into the Subversion repository? Stop the emails from coming?
If you look in the directory /group/halld/Repositories/svnroot/hooks there is a file called post-commit, with several backups. That is the file you need to change. If you look at it it is pretty clear what to do. People in the halld group should have privilege to modify it.
Note that you can "subscribe" to only a subset of check-ins as well, selection by directory.
How do I add a new author to the DocDB?
Any GlueXer with the writer privilege can add a new author. Go to https://halldweb.jlab.org/doc-private/DocDB/AuthorAddForm . (Thanks Zisis!)
How do I get write access to the GlueX GitHub repositories?
- To get an account go to https://github.com and "sign up". It is a typical web account creation activity. At this stage you are creating a personal account. The account you are creating has no affiliation with GlueX, Hall D, JLab, etc. As such, any email address is fine. The "Free" plan is fine as well. And obviously, if you already have GitHub account you can skip this step.
- After that you will have to join the "gluex" team under the "jeffersonlab" organization. To do that send an email to one of the Managers (listed below) with your GitHub username. They will send you an email invitation through the GitHub site. Once you accept the invitation, you will able to "push" changes to our repositories on GitHub.
- GlueX Team Managers
- Sean Dobbs
- Mark Ito
- David Lawrence
- Justin Stevens
- Alex Austregesilo
- GlueX Team Managers
How do join the JLab Slack channel?
To join, use this link. You must use your JLab email address when joining.
Building GlueX Software
How do I do a complete build of GlueX Software, starting from scratch?
See the instructions in "The gluex_install System" section of the Build Scripts document.
Where do I find version set files?
To get the version sets past and present, there are several options.
- Go the the webpage https://halldweb.jlab.org/halld_versions/ and click on the version set file of your choice. If you download the resulting page, you will get a local copy of the XML file (even though it looks like HTML).
- At JLab, from the command line, navigate to the directory /group/halld/www/halldweb/html/halld_versions . This is the directory pointed at from the URL above.
- Go to GitHub and browse the repository: https://github.com/jeffersonlab/halld_versions
- Clone the halld_versions repository from GitHub, for example
git clone https://github.com/jeffersonlab/halld_versions
Generally, only the master branch is used.
The latest versions are indicated by soft links. At present:
lrwxrwxrwx 1 gluex halld-2 15 Jul 31 10:40 version.xml -> version_4.15.0.xml
--marki (talk) 13:29, 19 February 2020 (EST)
What are "directory tags?"
Directory tags are attributes used to distinguish multiple instances of builds of a fixed version of a package. They get encoded in the directory name of the package build instance. For example, assume package Y (at version 2.0) depends on package X (at version 1.1). When a new version of package X comes out (version 1.2), package Y needs to be rebuilt, even though its version has not changed. The old package Y cannot be deleted; folks may be still be using it for their work with an older version set.
Old version set:
<package name="X" version="1.1" /> <package name="Y" version="2.0" />
with directory names "X-1.1" and "Y-2.0".
New version set:
<package name="X" version="1.2" /> <package name="Y" version="2.0" dirtag="X12"/>
with directory names "X-1.2" and "Y-2.0^X12".
Here the choice of "X12" as a directory tag is arbitrary. It has no functional significance. It only serves to distinguish the two instances of package Y, version 2.0.
How do I upgrade my $GLUEX_TOP?
New versions of the packages come out all of the time. Sometimes you have to upgrade.
1. Update your halld_versions directory.
cd /your/location/for/gluex_top/halld_versions git pull
This will update the default version (version.xml) to be linked to the latest version set.
2. Update your build_scripts directory.
cd /your/location/for/gluex_top/build_scripts # cd into build_scripts ./update_to_latest.sh # this must be executed from inside the build_scripts directory
If you don't see update_to_latest.sh in your build_scripts directory, checkout the master branch and do a git pull. The update_to_latest.sh should appear.
3. Repeat the gluex_install step
cd $GLUEX_TOP gxclean # clean your environment cd .. # go to the directory above $GLUEX_TOP /path/to/gluex_install/gluex_install.sh -r # re-run the gluex_install.sh script
Without the "-r" option, gluex_install.sh will refuse to do an install on top of a pre-existing gluex_top directory.
Also if you want a parallel build with, say, 12 parallel streams, set NTHREADS=12 in your environment.
If the above fails
There might be a gazillion of reasons. One of them could be missing system packages. First make sure you have the latest prereqs scripts in your gluex_install folder
cd /your/location/for/gluex_install # this is the folder above your gluex_top git pull
For the next step, most likely, you need to become root first
cd /your/location/for/gluex_install/ ./gluex_prereqs_centos_7.sh # install system prerequisites (CentOs 7 in this example)
JLab
How do I get a computer account at JLab?
You need to register as a JLab user. The registration form has a box for "I want an account" (or something like that). In any case registration must be completed before getting an account. Also you will need a sponsor. Your sponsor must be a member of JLab staff. The process starts with a link on this page
https://www.jlab.org/hr/jris/processing.html
How do I install a certificate in my browser so that I can view secure web sites at JLab (without creating a security exception)?
Go to https://pki.jlab.org/ , select the browser you are using, and follow the instructions.
I cannot log into the ifarm machines. How do I get access?
You have to be member of the "halld" Unix group (or an analogous Unix group associated with one of the other experimental halls).
How do I become a member of the "halld" Unix group?
See the JLab Knowledge Base for instructions and contact information for the Software/Computing Coordinator to request access.
How do I subscribe to a GlueX email list?
There is a wiki page listing email lists of interest to GlueX collaboarators. To subscribe to any of them, click on the appropriate "information page".
How do I get personal web space at JLab?
The standard /home/username/public_html has been deprecated at the Lab as of August 2011. Another system has been set up on the /userweb partition. There is a HOWTO that explains the procedure for getting space there.
What are the various types of public disk space available at JLab?
| Type | Directory | Size (TB) | Availability |
|---|---|---|---|
| scratch | /u/scratch | 23 | Lab-wide |
| group | /group/halld | 1.3 | Hall-D-only |
| work | /work/halld | 191 | Hall-D-only |
| volatile | /volatile/halld | 30-50 | Hall-D-only |
| cache | /cache/halld | 200-400 | Hall-D-only |
| osgpool | scosg16:/osgpool | 27 | Lab-wide |
--marki (talk) 11:30, 7 September 2018 (EDT)
What is the scratch disk?
The scratch disk is for temporary storage of files of all types. It is available from nearly all IT-Division-maintained nodes including jlabl?, ifarm?, and farm? nodes. It is also mounted on the gluon cluster.
Files left unaccessed on the scratch disk are deleted after sixty days automatically.
What is the difference between /u/scratch and /scratch
- On the jlab? nodes they are the same, the above mentioned network mounted scratch disk.
- On farm and ifarm /u/scratch is the networked mounted scratch disk (as it is on the jlab? nodes). /scratch is a locally mounted disk and therefore only visible to the local node.
The typical mistake one makes is to write stuff to /scratch/user when working on the ifarm and then look for those files in /scratch/user on a jlabl? node later. The files appear to have disappeared. Very confusing, no?
What is the Volatile disk?
The volatile disk is for temporary storage of large files, on the time scale of weeks. Find documentation here.
What is the Cache disk?
The cache disk is the disk area where files are staged from and staged to tape. Find documentation here.
How do I check how much space Hall D is using on the Lustre disks?
From a recent CCPR:
You can see the current lustre quota, and usage, for Hall D with the following:
> lfs quota -hg halld /lustre
Disk quotas for group halld (gid 267):
Filesystem used quota limit grace files quota limit grace
/lustre 103.6T 115T 120T - 7666476 0 0 -
Note that this shows the Hall D quota for all of the Lustre-based disks; the quota has no knowledge of cache/volatile/work.
Why does sudo not work for me?
At JLab, /apps/bin/sudo is configured for use by Computer Center personnel. It does not reference your local sudoers file. To use sudo locally, make sure you are using /usr/bin/sudo (i. e., check your PATH variable).
How do I kill all of my farm jobs?
jkill 0 will kill all farm jobs submitted by the logged in user, independent of the state of the jobs.
How to access RCDB with python (e.g. plot accumulated triggers)
- On most machines at JLab
- Source /group/halld/Software/build_scripts/gluex_env_jlab.csh
- Use /apps/anaconda/PRO/bin/python2
- Example: > python2 plot_rcdb3.py # run script that plots accumulated triggers over the run
How do I get a Scientific Computing Certificate?
/site/bin/jcert -create
if you are getting one for the first time, or
/site/bin/jcert -replace
if you have an expired certificate and want to get a fresh one.
For more details see SciComp's network certificate page.
The computer center maintains a page on that here. You need to scroll down to see the table.
How do I run a remote desktop session on a JLab machine from my Windows machine?
See HOWTO establish a remote desktop on the ifarm starting from Windows.
How do I connect to the JLab VPN?
Using the JLab VPN makes the network on your local machine, located outside the lab, appear as if it is inside the JLab firewall. This gives you direct access to on-site resources (e.g., ssh ifarm) and subjects you to on-site restrictions (e.g., blocked webpages).
The Computer Center maintains a webpage with links for downloading the VPN client for Windows, Mac, and Linux.
There are no instructions for installation and execution on Linux on the webpage, but this page from UC Irvine gives instructions that are sufficient.
Remember:
- You want to connect your VPN client to vpn.jlab.org.
- You have to use your MFA method to login. Your CUE password will not work.
--marki (talk) 12:13, 2 October 2020 (EDT)
Is there a way to comment on a ServiceNow incident opened by another user?
Besides the servicenow IT group, only the requestor, affected user or someone on the watchlist can add comments to incidents.
The affected user or requestor should be able to add you to the watchlist which will allow you to add comments to a particular incident.
Farm Jobs
How do farm jobs handle input files requested in the job specification?
There is documentation here.
By default, Auger will not copy the files to the local farm node disk when the input files are from /mss (only a link will be made), and files from other location (/volatile, /work, /home etc.) will be copied to local disk. The idea is that files stored on /cache tend to be large and can be streamed. There is an attribute (copyOption="link or copy", see the documentation) so the user can specify copying or linking of the file.
-- Y. Chen, July 23, 2020
What is the relationship between slurm jobs and auger jobs?
Auger jobs are a subset of Slurm jobs, i.e. Auger ultimately submits jobs to Slurm, which runs them on the Farm. If you're not using the features of Auger (which I believe have mostly to do with stage-in/out) or SWIF2, which is intended to replace it, you can submit jobs directly to Slurm.
-- Helpdesk, October 26, 2021
Where do I find a list of valid PROJECT fields for farm jobs?
The fields are listed here.
-- Helpdesk, October 27, 2021
Is there an Auger/Slurm FAQ page somewhere?
Yes there is. Find it here.
-- Helpdesk, October 27, 2021
What is this sbatch error that I am getting?
If you are getting an error like
sbatch: error: Batch job submission failed: Invalid account or account/partition combination specified
one possibility is that you have not been added to one of the Slurm accounts. You can check for your username on this webpage. If you need to be added, please contact the Hall D Computing Coordinator. There are instructions for them to add you on this page.
--marki (talk) 09:08, 7 March 2022 (EST)
SWIF
How do I resolve problem jobs so I can start the workflow going again?
When you have hit the error limit, SWIF will stop submitting jobs from your workflow. By default, there is no limit set. If you have set a limit and have reached the limit, you can use 'modify-jobs' if you want to retry them with altered cores/time/ram/disk. If you simply want to try again, use 'retry-jobs'. To just fail them, use 'abandon-jobs'.
If you want to put off thinking about it, you can up the error threshold to, say, 1000 by using 'swif run -workflow <> -errorlimit 1000'.
Suppose a successful job, upon scrutiny later, looks suspicious and I want to re-run it. Is there a way to to this?
You can use the undocumented '-resurrect' option of 'swif retry-jobs' then specify which jobs to resurrect either by name or id. You can also specify job names and ids by pattern, but you need to make sure the pattern isn't too broad because this command will search all jobs, not just ones that were successful.
What do I have to know when transitioning from SWIF to SWIF2?
The issues are explained on this wiki page by Alex Austregesilo.
--marki (talk) 14:27, 1 March 2022 (EST)
JLab Administrative Topics
How do I check on the status of my JLab-mandated training?
Use this link
How do I find a list of JLab users that I sponsor?
Log in here: https://mis.jlab.org/mis/apps/training/ejta/index.cfm
Your list should be under "My direct reports"
--marki (talk) 16:20, 27 February 2020 (EST)
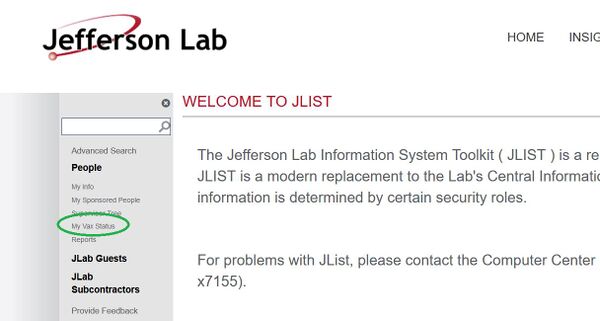
I work at JLab. How to I check whether the Lab has received my COVID vaccination status?
If you log into JList (you have to be inside the firewall), you can find the link to see your vaccination status under the “People” menu:
halld_recon
How do I get debug symbols in my halld_recon binary?
Debug symbols are put in by default by the SBMS system. You don't have to do anything.
How do I create EVIO data from an HDDM file produced by HDGeant?
hd_ana -PPLUGINS=rawevent file.hddm
halld_sim
How do I get debug symbols in my halld_sim binary?
Debug symbols are put in by default by the SBMS system. You don't have to do anything.
When unstable particles appear in the simulation, which ones are decayed by the event generator versus hdgeant/hdgeant4?
In general, the choice is up to you. If you are writing an event generator and you want to produce an unstable particle like a τ- lepton or an η meson, you can either convert it immediately into the daughter particles and pass those to hdgeant/hdgeant4 for tracking, or you can pass the unstable particle directly to hdgeant/hdgeant4 and let it decay in the simulation. In order for hdgeant or hdgeant4 to handle the decay, the mother particle needs to be included in the list of known particles to the underlying Geant library. The known particle lists for the two Geant versions are similar, but not identical. See the following references for details.
- Geant3 Reference Manual, see Table CONS-301 on page 58-59.
- Geant4 User's Guide, separate tables for leptons, mesons, baryons, ions, and others.
Unstable particles that appear in these lists are those that the Geant/Geant4 library has some knowledge of, and can generate the major decay modes with the correct branching ratios. The fact that these libraries can simulate the in-flight decays of these particles does not mean that it should be done that way. There are limitations to the decay models employed in these simulation libraries, particularly the fact that the daughter particle momenta are distributed uniformly in phase space. For example, if you inject a bunch of ω(783) mesons into hdgeant or hdgeant4 and then generate a Dalitz plot of their 3-pion decays, you will see that the interior of the Dalitz ellipse is uniformly populated rather than weighted by the physical amplitude for isoscalar,vector -> 3π decays. One way to address this would be to generate the decays instead within the event generator, using the proper physics amplitude. Then hdgeant/hdgeant4 would only see the 3 pions from omega decay and would propagate/decay those according to their lifetimes and branching fractions. The more standard approach to this problem in GlueX analyses would be to generate all of the hadronic decays using a uniform phase-space distribution, and then apply the physical decay distribution later on in the analysis as a factor in the PWA amplitude. However, that is beyond the scope of this FAQ. For the present purposes, you can either embrace phase-space decay distributions and let hdgeant/hdgeant4 perform the decays, or else take control of the decays directly in the event generator and make the decay distributions as realistic as you wish. In general, I recommend the following rule of thumb as a best practice for GlueX simulations.
If the unstable particle has a sufficiently long lifetime that it can travel a measurable distance (>0.1 mm) in the detector before decaying, the decay should be handled by hdgeant/hdgeant4. If on the other hand, the unstable particle lifetime is too short to allow its decay vertex to be distinguished from the primary event vertex then the decays should be handled by the event generator. This second rule gives the experimenter more flexibility in terms of which decay branches are selected for simulation, and saves time that otherwise would be spent in the simulation following products of decay branches that are not the focus of the study.
"But," someone asks, "I don't want to write my own generator. I want to use bggen (pythia) or genr8, or some other channel-specific generator that I found in the sim-recon codebase. What happens then?"
The answer depends on the specific event generator tool you are using, but in general they tend to conform to the best-practice rule given above. In particular, bggen only generates the following unstable objects as final state particles.
- AntiLambda
- AntiNeutron
- AntiSigma+
- AntiSigma-
- AntiSigma0
- AntiXi+
- AntiXi0"
- Eta
- K+
- K-
- KLong
- KShort
- Lambda
- Muon+
- Muon-
- Neutron
- Pi+
- Pi-
- Pi0
- Sigma+
- Sigma-
- Sigma0
- Xi-
- Xi0
You can verify that all of the unstable particles in this list are known particle types to both hdgeant and hdgeant4. Any unstable objects not in this list, that appear in bggen as products from the primary interaction vertex, are tagged in the generated output file as intermediate resonances, not final state particles. When reading from the bggen event source, the hdgeant/hdgeant4 simulation tracks only those that are tagged as final state particles, ignoring the intermediate nodes in the decay tree. This way there is no double-counting of parent and daughter particles in the simulation, yet the record of which final-state particles conceptually belong to which parent resonances is still available in the simulation output file in case the user wants to check it.
CCDB
What do I need to specify to get the correct constants from the CCDB?
There are four items to specify:
- run number
- database source
- variation
- calibration time
1. Run Number
Each application has a built-in mechanism for choosing the run number. See the GitHub wiki documentation for the API used by the application.
2. Database Source
The connection string specifies the database source.
- Syntax: dialect://username:password@host:port/database
- Examples:
- mysql://user_name:password@host:port/database
- sqlite:///path_to_file
- Reference: GitHub Wiki Documentation: Concepts
3,4. Variation Choice and Calibration Time
For JANA, the JANA_CALIB_CONTEXT variable contains information about variation choice and calibration time.
- Example:
- JANA_CALIB_CONTEXT="variation=john calibtime=2014-05"
- Reference: Specifying_Alternate_Versions_of_Calibration_Constants_in_JANA
How do I use a CCDB SQLite file?
There are instructions on this wiki page.
I am getting a disk I/O error reading an SQLite version of the CCDB. What is wrong?
If your error looks like
> ccdb ls CCDB provider unable to connect to sqlite:////work/halld/user/ccdb.sqlite. Aborting command. Exception details: (sqlite3.OperationalError) disk I/O error [SQL: u'SELECT "schemaVersions"."schemaVersion" AS "schemaVersions_schemaVersion", "schemaVersions".id AS "schemaVersions_id" \nFROM "schemaVersions"\n LIMIT ? OFFSET ?'] [parameters: (1, 0)]
it is likely you are trying to use a SQLite file from a Lustre-based disk system. SQLite is incompatible with Lustre. You must move your file to a different, non-Lustre file system.
Simulation
How are random triggers staged for the simulation?
See this wiki page.
Git
Where can I get help with Git?
See Git Help Resources on this wiki.
What are the basic git commands that I need to know?
- git help: get a help message (e. g., git help clone)
- git clone: get a complete copy of a repository
- git status: show the status of your files (changed, added to index, etc.)
- git checkout: change to another branch
- git add: add modified files to the index
- git commit: commit files in the index to your repository
- git branch: show, create, delete branches
- git tag: show, create, delete tags
- git fetch: get remote repository information, do not change files
- git pull: get changed files from remote repository
- git push: write changed files to remote repository
What happens when I do a git clone?
When you clone a repository to your local disk, you get not only get the main line of code (in SVN: the trunk, in Git: the master branch), but all branches and tags and the history of all of the above. The directory that is created contains the actual files being controlled consistent with the master branch. In the top level directory there is a hidden directory, .git, that contains all of the versioning information. There are no .git directories in the subdirectories. You are meant to work with the files you see. You do not checkout the code to a separate directory as you do with SVN or CVS. The repository and the working files co-exist in the same tree.
What is a remote repository?
A remote repository is any repository outside of your repository that your repository is aware of. A network of remote repositories will have a common ancestor. When you clone a repository, your new repository is automatically aware if one remote repository: the repository from which it was cloned. The remote repository is called "origin" on your local repository by default. Remote repositories can be added and removed. They can be pushed to or pulled from. To see your remote repositories do a "git remote".
What is a tracking branch?
If you're on a tracking branch and type git push, Git automatically knows which server and branch to push to. Also, running git pull while on one of these branches fetches all the remote references and then automatically merges in the corresponding remote branch.
From http://stackoverflow.com/questions/4693588/git-what-is-a-tracking-branch
How do I create a tracking branch?
When branches are created using the --track option, they will be set up linked to the remote branch. For example, if you wanted to create a new branch from the master branch from the origin remote, using this would set it up so it would pull from the remote and branch automatically:
$ git branch --track feature1 origin/master
Branch feature1 set up to track remote branch refs/remotes/origin/master.
From http://gitready.com/beginner/2009/03/09/remote-tracking-branches.html
How do I make an existing branch track a remote branch?
As of Git 1.8.0:
git branch -u upstream/foo
Or, if local branch foo is not the current branch:
git branch -u upstream/foo foo
Or, if you like to type longer commands, these are equivalent to the above two:
git branch --set-upstream-to=upstream/foo
git branch --set-upstream-to=upstream/foo foo
As of Git 1.7.0:
git branch --set-upstream foo upstream/foo
All of the above commands will cause local branch foo to track remote branch foo from remote upstream. The old (1.7.x) syntax is deprecated in favor of the new (1.8+) syntax. The new syntax is intended to be more intuitive and easier to remember.
From http://stackoverflow.com/questions/520650/make-an-existing-git-branch-track-a-remote-branch
What is a topic branch?
A topic branch is a branch that is created, and possibly shared, to work on some particular issue. The name of the branch should be descriptive of the issue, of course. The concept emphasizes separation of work on one issue from work on others. One may have several topic branches going at the same time. The word "topic" is descriptive and has no operational significance.
What is the difference between Git and GitHub?
Git is the underlying software package that performs the direct operations on Git repositories. It is open source. As a software project, it is completely independent of GitHub. One can profitably run a project under Git without ever using a web browser.
GitHub is a commercial site that is in the business of hosting Git repositories and providing web tools for working with them. Jefferson Lab has a contract with GitHub to host repositories for doing Lab-related work. Public repositories are free; private repositories can potentially incur charges.
How do I get privilege to create pull requests on GitHub?
Find the instructions here.
How do I authenticate to GitHub from the command line?
There are instructions on the GitHub site for this. For most folks HTTPS and SSH are the two relevant methods.
The method you use to authenticate has to correspond to how you cloned the repository originally. If it was cloned with "https://" as the name of the repo, you have to use the personal access token. If you cloned using "git@github.com:", then the ssh key is used.
Author's note:
- For personal use, I always checkout out using ssh. So I had to load my public ssh key to GitHub. Using an ssh-agent as a component of this method means that I never have to enter a password to GitHub for these personally cloned repos.
- On the other hand, I often run across repos that have been cloned for public use (e.g., the public version of build scripts, or halld_recon in the nightly build). If changes have to be pushed back to GitHub, I then have to use my personal access token.
--marki (talk) 13:14, 1 February 2022 (EST)
Why is there no "git pull-request" command?
Generically, a pull request is any communication from one developer to another asking that changes on the one's branch be incorporated on the others remote branch via a "git pull". As your question implies, there is no native Git command for doing this.
On GitHub, there is a site-specific set of tools (web pages and web forms) for issuing pull requests which includes a conversation between developers on the proposed change. These tools work only for the case when the source and the target branch are both hosted on GitHub. The repositories need not be owned by the same account.
How are we notified of pull requests?
Those interested in acting on pull requests should "watch" the appropriate repository on GitHub. Go to settings/notification to configure how you are notified (email, website, or both). Then go to the repository page to mark the repository as being watched/not-watched/ignored by you.
What other notifications are available?
GitHub provides a facility for notification of several types of activities on the site. You can choose to view your notifications through the notifications inbox at https://github.com/notifications or through your email. For a description see their Setting up notifications page.
What is the difference between a clone and a fork?
A fork is a clone from a GitHub-hosted repository to another GitHub-hosted repository. It is a GitHub-specific term.
What happens to forks when a repository is deleted or changes visibility?
When you delete a public repository, one of the existing public forks is chosen to be the new parent repository. All other repositories are forked off of this new parent and subsequent pull requests go to this new parent.
What happens when I checkout another branch when there are uncommitted changes on the current branch?
If the new branch contains edits that are different from the current branch for that particular changed file, then it will not allow you to switch branches until the change is committed or stashed. If the changed file is the same on both branches (that is, the committed version of that file), then you can switch freely.
I cannot clone a GitHub repository on the Gluon Cluster. What is the trick?
setenv HTTPS_PROXY https://jprox:8082
or
export HTTPS_PROXY=https://jprox:8082
I am getting a "403 Forbidden" error when trying to push to a GitHub repository from JLab. What is wrong?
You might be using an old version of git. On the JLab CUE or on the Gluon Cluster, use the version of git from /apps/bin and not the one in /usr/bin. (Version 1.7.1 has given problems; version 1.7.4.5 seems OK. See https://help.github.com/articles/https-cloning-errors/). In addition the version of /apps/bin/git on the CentOS 6.2 farm nodes (e. g., ifarm62) gives error messages. It is not known if commands are executing correctly or not.
After a branch is deleted on GitHub, I still see it when I do a git branch -r on my local repository, even after a git fetch. How do I get rid of them?
git remote prune origin will remove all such stale branches.
From http://stackoverflow.com/questions/5094293/git-remote-branch-deleted-but-still-appears-in-branch-a
How do I get to the compare view on GitHub?
To get to the compare view, append /compare to your repository's path.
Tags have changed on a remote repository. How do I update them on my local repository?
On the local clone:
git fetch --prune --tags
I am getting SSL errors when trying to access GitHub from JLab. What is wrong?
For example, you might see the error:
Peer certificate cannot be authenticated with known CA certificates.
or
Peer's certificate issuer has been marked as not trusted by the user.
In late 2016 through early 2017, after JLab enacted stricter security measures for encrypted web traffic (e.g. HTTPS), users had to reference a JLab security certificate in their git configuration when authenticating to GitHub. After that period, the reference is not only unnecessary, but prevents git from working with GitHub repositories using https.
To get rid of the reference remove the "sslCAInfo" line from your ~/.gitconfig file.
How do I use secure shell (ssh) to access GitHub?
You can clone a repository, for example halld_recon, by typing
git clone ssh://git@github.com/jeffersonlab/halld_recon
or, since ssh is the default transport mechanism
git clone git@github.com:jeffersonlab/halld_recon
at the command line. Note that you have to specify user "git" in the URL. This will not work unless you have previously added your public ssh key to your GitHub account. To add your key:
- Log into GitHub
- Navigate to "settings" (click on far upper right icon, select "settings")
- Click on "SSH and GPG keys" in the left column.
- Click on "New SSH key" (near the upper right)
- Enter a title (reminder about where the key is from) and your public key.
- Click on "Add SSH key"
For any repository you clone using ssh, you use your ssh passphrase to authenticate. This allows you to avoid the long, un-memorizable personal access token.
If you run an ssh-agent that provides your passphrase, you can operate password free.
What if after this I get the "X11 forwarding request failed on channel 0" message?
That does not affect the success of your command. To get rid of it create a file ~/.ssh/config that contains
Host github.com
ForwardX11 no
with only user read/write permissions. After that you will not get the message.
How do I restart the automatic pull-request test?
From the main GitHub page:
Settings -> Webhooks -> [select the script, it's the only one]
Scroll down to "Recent Deliveries", and if you click on the hashes, it gives you details about the messages that were recently sent to the script. If you look at those, and pick the one corresponding to the pull request you are interested in, with an "action" of "opened", you can click the "Redeliver" button to trigger the action again.
You can go to the GlueX "team" page and click on the "Repositories" link, or more directly, click here.
How do I get notified of changes in a repository on GitHub?
See the Configuring notifications page on the GitHub site.
ROOT
How to use ProofLite
A good place to start learning about how to use ProofLite is USING ProofLite WITH DSELECTOR
How do I get rid of zombie ProofLite servers?
We were having a problem with zombie proofserv.exe processes left behind on our cluster nodes after the jobs completed, they were not cleaned up by slurm. Every so often, one of many worker threads would fail to initialize properly, as seen in the note on how many threads had gone parallel. The job output was unaffected, although presumably it took fractionally longer than it should have done, but after hundreds of DSelector jobs had been run, there were a large number of zombie processes left behind.
It turned out to be sort of a network issue, which caused TProof to timeout (after only 5 seconds) on some of the forked threads. Since it timed out, it skipped that thread and did not destroy it when ROOT exited. Setting a longer timeout solved the problem. This can be done by appending the following line to ${HOME}/.rootrc
ProofLite.StartupTimeOut 1800
which gives a very generous timeout allowance of 30 minutes.
--nsjarvis 19 August 2020
What do I have be careful of when creating ROOT histograms and trees with JANA?
See this wiki page: Locking in JANA.
--marki (talk) 10:14, 1 February 2022 (EST) (thanks to Sdobbs for pointing this out recently)